Automated Monitoring of CAR-T Cells in a Rocking Motion Bioreactor
Jérémie Barbau, Laurent Desmecht, Serge Jooris.
Ovizio Imaging Systems SA/NV, Brussels, Belgium
Introduction
Chimeric Antigen Receptor (CAR) T-cell therapies are showing high response rates in patients worldwide, resulting in two approved products by the US Food and Drug Administration in 2017. T-cells from a patient are removed from the blood and engineered to express the CAR to reprogram the T-cells to target patients tumoral cells. (1)
In these autologous therapies, the challenges lay in the inherent variability in starting materials and the goal of maximizing product consistency while producing a safe, pure and potent product. Cell counting is one of the most fundamental metrics of it. With the development of Cell Therapy Products (CTPs), there is an increased need for robust and validated measurements for cell characterization to enable manufacturing control and absafe/high-quality product suitable to be released to the patients. (2)
In CAR-T cell manufacturing, each handling or addition of reagents to the cell preparation generates a risk for error and for contamination that can possibly lead to the loss of a production run. A reliable solution consists of removal of open handling and implementing closed culture systems, where the cell manufacturing takes place in bags with closed tubing pathways and connections, maintaining a sterile environment. Ovizio’s patented technology, Double Differential Digital Holographic Microscopy (D3HM), is a quantitative imaging technique that allows cell monitoring in a continuous, automated and label-free set-up. No need for sampling (eliminating the risk of contamination), staining, and waiting for results generated by an off-line counter or analyzer; therefore, results are available in nearly real-time and continuous spanning the length of the culture. The platform generates a holographic fingerprint based on 70 parameters for every cell that is imaged, and feeds this data to a machine learning platform that can be trained to identify cells and discriminate between cell types. Fast and accurate, the algorithms automatically discriminate living from dead cells, count and give access to in-depth quality attributes and dynamic properties of your samples, and may also provide additional information on a single cell level.
In this application note, the use of the iLine F, manufactured by Ovizio Imaging Systems, for the monitoring of T-cells as they grow in a rocking motion bioreactor is described. The iLine F delivers reliable measurements of viability, cell density, diameter evolution and specific cellular critical quality attributes, allowing for real time in-process controls. Typically, the iLine F (i) provides a continuous monitoring of T cells culture in wave bags, (ii) counts and discriminates the viability of the T cells, (iii) gives strongly comparable Total Cell Density (TCD) and Viable Cell Density (VCD) with an off-line reference counting method (R2 <0.95) and, (iv) tracks small phenotype changes allowing for a T lymphocytes classification (subsets, differentiation states (still under evaluation)).
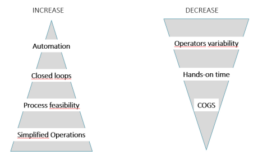
Figure 1. The needs and expectations associated with manufacturing of Cell Therapy Products.
Ovizio’s technology
Digital Holographic Microscopy is the scattered light beam from an illuminated object that interferes with a reference beam on a CCD camera allowing for a 3D numerical reconstruction of that object. Double Differential Digital Holography (D3HM) (Figure 2) is an evolution of this base technology that enables the use of a partial coherent light source (low power, non-invasive LED light, e.g.) resulting in improved image quality as well as an important size reduction of the instruments. A patented method is used to discriminate live from dead cells: a living cell containing cytoplasm is spherical and will create an out of focus light cone, whereas a dead cell losing membrane integrity and cytoplasm will diffuse light (Figure 2). One of the most important features of holographic microscopy is the capability of refocusing out of focus objects post acquisition. This makes the platform extremely suited of tracking objects in suspension.
The iLine F microscope acquires both intensity and quantitative phase contrast information of a microscopic sample allowing the automated extraction of 70 parameters for each object captured- the object being a cell (dead or alive).
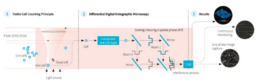
Figure 2. Ovizio’s Double Differential Holographic Imaging Technology. Live or dead cells are the objects (1) that pass
through the light scattering and interference process (2) resulting in the 3D reconstruction and quantitative phase data (3).
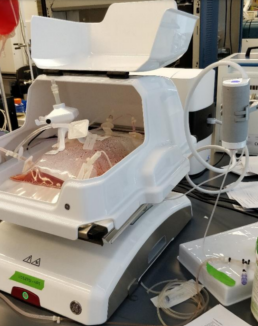
The iLine F can perform continuous suspension cell monitoring in rocking motion (wave) type bioreactors via an adapted setup (Figure 5). The disposable BioConnect sampling probe in combination with a reusable pump is a closed loop that pumps cells out of the wave bag through a flow cell that inserts into the iLine F microscope, holographic data is then acquired, and cells flow back into the wave bag. The holograms are analyzed by OsOne, the monitoring software part of the iLine F microscope and a holographic fingerprint is generated for every cell found within the hologram. The fingerprint is then used by a machine learning platform trained to identify, count and analyze the sample. Viability, transfection kinetics and other cellular parameters can be detected by the platform.

Figure 3. Machine learning principle.
The system can be trained to discriminate and count different subpopulations by observing pure populations (100% population “A”) and then observing known mixtures of that same subpopulation in incremental steps (90%, 80%, 70%, … 10%, e.g.). The system can either automatically, or assisted by supervised learning methods, define which parameters are best to discriminate the sub populations.
Materials
- iLine F D3HM device
- Single use BioConnect monitoring probe
- Single use adapter for disposable bags
- Reusable pump engine
- OsOne Software version 5.12
- GE Healthcare Xuri Cell Expansion System W25
- GE Healthcare Cellbag 2L DOOPT II, pHOPT and Perfusion (reference 29-1054-98)
- An offline reference counting method making use of fluorescent dyes
Methods
The BioConnect, Ovizio’s disposable sampling probe for bioreactor monitoring is directly connected to a disposable cell culture bag. Two silicone tubes that terminate with C-flex and male Luer locks are used to connect to the wave bag, either by welding or under sterile conditions (within a LAF hood). The BioConnect was sterilized in an autoclave for 20 minutes at 120°C and connected aseptically to the cell
culture bag using welding.
The inlet tubing of the BioConnect is connected to the harvest line in order to take advantage of the dip tubing in the cell culture bag while the outlet can be connected to the sampling port. Sampling was still possible by adding a single use (sterilized by autoclave), T-shaped adapter between the sample port and the BioConnect.
Figure 4. Modified BioConnect.
Figure 5. Complete wave bioreactor setup.
Once the assembly finalized, OsOne software was launched and a wizard designed to guide users through the selections of the proper cell type, and the number of data points to be acquired during the run, was completed.
Figure 6. OsOne software.
OsOne is an all-in-one software that:
- controls the iLine F microscope
- acquires holographic fingerprints of each individual cell within the wave bag
- computes the results (data analysis)
Cell density, viability and the growth curves (viable and total cell density) are displayed on the screen for easy viewing while operating. The bioreactor was inoculated with 1×106 viable T cells/ml. For this experiment, the iLine F has been set to acquire holograms every 2 minutes and computes a data point every hour. At low cell densities the system automatically doubles the number of images captured to increase statistical relevance. For every single cell imaged by the platform, 70 parameters are recorded and available for additional post processing if required.
RESULTS
OsOne plots results graphically on the screen in nearly real-time (Figure 6). During the complete run, images of the cells are acquired every two minutes. Data can be exported in multiple formats and archived for future analyses.
During the experiment data was acquired for up to 4 days continuously by the iLine F microscope and was compared to results generated by the off-line reference instrument. On average, two manual samples were acquired for off-line analyses per day.

Figure 7. Total Cell Density and Viable Cell Density compared to the off-line reference method.
Figure 7 illustrates Total Cell Density (TCD) and the Viable Cell Density (VCD) compared to the reference method used. After plotting the data points, the correlation coefficient (R2 ) was calculated and resulted in a value of 0.96. This measurement represents the actual deviation between the compared methods with respect to a linear correlation assumption, that is, if cell densities grow by a factor of two, then both methods are expected to report a factor of two. A perfect correlation results in an R2 value of 1, though, it does not indicate to whether
the absolute measurements are the same.
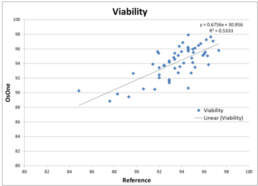
Figure 8. Viability compared to the off-line reference method.
Figure 8 illustrates Viability as provided by the iLine F compared to the reference method used. After plotting the data points, the correlation coefficient (R2 ) was calculated and resulted in a value of 0.53. This value is lower compared to the value obtained when comparing TCD and VCD with the reference method. It can be explained by the fact that viability stays around 95% during the course of the culture. Indeed very few data points can be found below 90%. It results in the values being part of a cloud of points instead of being distributed along a line. As a consequence, fitting a regression line is much more complicated hence the low value for the correlation coefficient. However, the discrepancies between the values generated by the iLine F and the reference method are minor: mostly within 5% of variability.

Figure 9. Total Cell Density and Viable Cell Density compared to the off-line reference method within a 95% confidence
interval.
A 95% confidence interval has been fitted on the Total Cell Density and the Viable Cell Density, showing that most of the values are within this confidence interval.

Figure 10. Donor-related variation shows that there is no influence from the donor on the quality of the measurement.
Donor-related variations have been investigated. Figure 10 gives an overview of 3 different donors. Differences in the cell growth pattern can be seen while the correlation between the data provided by the iLine F and the reference method remain very good for each donor. It shows that the system is able to produce reliable data, independently of the donor.
It has to be noted that at the start of the run, there are less cells and thus more empty images, therefore, we observed some variability in the results when compared to the first three off-line data points. After 24 hours the correlations improved as the density increased with culture growth. Improvements have been made in the OsOne software to acquire more images at low densities to mitigate this limitation.
CONCLUSION
This study illustrates the robustness and reliability of Ovizio’s label-free approach for T-cell based expansion in process development or manufacturing environment. We have addressed the need to understand the biological basis for cell counting and characterization, especially when a subpopulation of cells is hypothesized to correlate with a clinical outcome.
The iLine F, in-line technology combined with the machine learning based analysis software OsOne, delivers results that are nearly equivalent to traditional off-line methods. Moreover, it provides additional information on a single cell level for continuous monitoring of CAR-T cell cultures. As the system is automated, there is also no need for an operator to sample the cell culture bag, thus eliminating operator-related variability and reducing operational costs.
The iLine F and adapted BioConnect disposables can be used as a Process Analytical Tool in a closed manufacturing environment with improved product characterization maintaining sterility throughout the cell expansion process. The instrument can be connected to the manufacturing control environment via OPC and is CFR 21 part 11 compliant. Each run is archived, and data can be accessed for analyses or regulatory purposes.
REFERENCES
- Towards a commercial process for the manufacture of genetically modified T cells for therapy, A D Kaiser, M Assenmacher, B Schröder, M Meyer, R Orentas, U Bethke & B Dropulic, Cancer Gene Therapy volume 22, pages 72–78 (2015)
- Manufacturing Cell Therapies: The Paradigm Shift in Health Care of This Century, Rachel Haddock, MS, GlaxoSmithKline; Sheng Lin-Gibson, PhD, National Institute of Standards and Technology; Nadya Lumelsky, PhD, National Institute of Dental and Craniofacial Research, National Institutes of Health; Richard McFarland, PhD, MD, Advanced Regenerative Manufacturing Institute; Krishnendu Roy, PhD, Georgia Institute of Technology; Krishanu Saha, PhD, University of Wisconsin– Madison; Jiwen Zhang, PhD, GE Healthcare; Claudia Zylberberg, PhD, Akron Biotech, June 23, 2017
Real-time and label-free automated cell monitoring: Shed light on your cells
Any lab faces bottleneck challenges when scaling up activities.
When scaling up your lab activities, bottlenecks can occur in multiple phases of handling cells in culture vessels:
- manual sampling
- sample preparation
- staining
- feed to offline instrument
- readout by trained operator
- disposal of sample

On-line and dye-free automated cell monitoring: bring automation to cell expansion
Ovizio’s value proposition:
- A closed loop cell monitoring system decreases biosafety risks.
- Real-time cell culture analysis increases quality control and improves proces controls.
- Automation and dye-free analysis of cell culture decreases development costs and operating costs.
The connection to our cell culture analyzer is bioreactor agnostic.
Our system can connect with many types of bioreactors:
- small scale bioreactors
- single use bioreactors
- rocking motion bioreactors
- benchtop bioreactors
- stainless steel bioreactors
Different bioreactor connection types are supported:
- c-flex or pvc welding
- luer lock connection
- novaseptum sterile connection

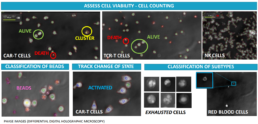
3D image based automated cell monitoring provides immune cell fingerprints.
Our cell culture analysis system uses differential digital holographic microscopy for:
- assessing cell viability
- cell counting
- classification of beads
- tracking changes of state
- classification of subtypes
Automated, closed-loop, in-line monitoring of CAR-T cells in a production process
Double Differential Digital Holographic Microscopy (D3HM)
Cell-based technology is a fundamental pillar of modern biotechnology. Cell counting is one of the most fundamental metrics of it. With the development of Cell Therapy Products (CTPs), there is an increased need for robust and validated measurements for cell characterization to enable manufacturing control and a safe/high-quality product released to the patients .
Our company has developed an in-line, automated microscope to monitor in real-time the suspension culture in a bioreactor. Its versatility makes it compatible with off-the-shelf stirred tanks, wave bags and others. The cell characterization and quantification are based on OVIZIO’s patented technology: Double Differential Digital Holographic Microscopy (D3HM). The microscope generates a holographic fingerprint based on 70 parameters for every cell that is imaged and feeds to a machine learning platform. Fast and accurate, the algorithms automatically discriminate living from dead cells, count and give access to in-depth quality attributes and dynamic properties of your samples, and may also provide additional information on a single cell level.
In this study we show that our in-line microscope delivers a continuous monitoring of T cells culture in wave bags, counts and discriminate the viability of the T cells, gives strongly comparable Total Cell Density (TCD) and Viable Cell Density (VCD) with an off-line reference counting method (0,93> R2 >0.99) and, tracks small phenotype changes allowing for a T lymphocytes classification (subsets, differentiation states (still under evaluation)).
This study illustrates the robustness and reliability of OVIZIO’s label-free approach for T-cell based expansion in process development or
manufacturing environment. We have addressed the need to understand the biological basis for cell counting, especially when a subpopulation of cells is hypothesized to correlate with a clinical outcome.
Online monitoring of the baculovirus expression vector system in bioreactors
Background
The increasing threat of pandemics due to the appearance of new viral diseases and the increased spread of existing viruses globally urges the development of a flexible vaccine production platform. Virus like particles (VLPs) form a promising new vaccine concept and can be produced using the baculovirus expression vector system and insect cells. Online determination of the viable cell density and state of infection can help in determining the optimal parameters for the production process.
The iLine F microscope system continuously pumps suspension cells in culture through a measuring chamber in a closed-loop system and makes holographic images of the cells resulting in up to 70 parameters per individual cell. Thus, it can accurately determine cell concentration and cell shape. The next step is to study whether it can also measure the infection state of the cell.
Objective
Evaluate online 3D holographic microscopy (iLine F) to determine the fraction of infected cells.
Results
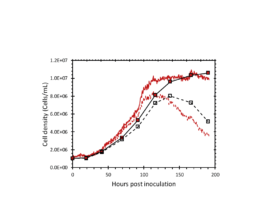
Figure 2: The total and viable cell density determined by the iLine F (red lines) and offline counts (square markers).
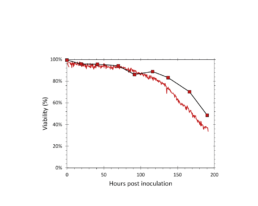
Figure 3: The viability determined by the iLine F (solid red) and trypan blue staining (square markers).
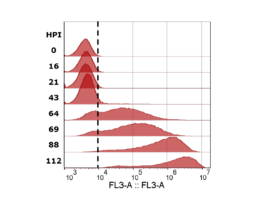
Figure 4: mCherry fluorescence measured with flow cytometer. Cells with values higher than the gate (dotted black line) were determined to be infected. Sample timepoints are given in hours post infection (HPI).
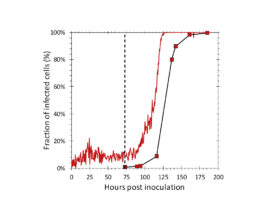
Figure 5: Fraction of infected cells determined by the iLine F (solid red) and the flow cytometer (square markers). AcBAC-PH-mCherry (MOI=0.1) was added to the reactor at 73 hours post inoculation (dotted black line).
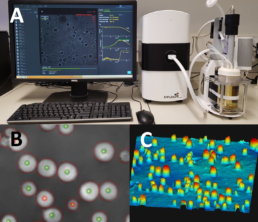
Figure 1. A: A single use bioreactor setup with the iLine F monitoring system. B: The cell segmentation and live dead determination performed by the microscope (green markers: live, red markers: dead). C: The 3D image created by the microscope.
Methods
• ExpiSf cells (Thermo Fisher) were cultured in ExpiSf chemically defined medium (Gibco)
• AcMNPVs: AcBAC-PH-HisGFP, AcBAC-PH-mCherry
• Applikon MiniBio reactors (400mL working volume, DO=30%, T=27⁰C)
• iLine F microscope system (OVIZIO)
• TC20 automated cell counter (Bio-Rad)
• Cells were infected at 3-4×10^6 cells/mL with a multiplicity of infection of 0.1 (TCID/cell)
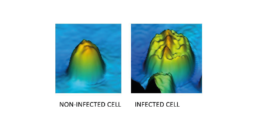
Figure 6: The 3D image made by the iLine F of a non-infected and infected Sf9 cell.
The iLine F showed a cell density and viability trend similar to the offline measurements. Figure 5 shows that the iLine F was able to detect baculoviral infection earlier than the offline flow cytometer method. Since mCherry was expressed behind the very late polyhedrin promoter (active at ~18 hours post infection) the flow cytometer method shown in Figure 4 and 5 only detected cells that were in the late stage of infection. Online holographic microscopy can capture changes directly as they occur on the cellular level, eliminating the need of offline sample preparation and reporter proteins such as mCherry. Detection is based on a combination of parameters generated by the microscope. A training of the algorithm was necessary (data not shown). With increasing amounts of data the algorithm can be further optimized and also used to re-analyze old data sets.
Conclusion
-
The iLine F microscope system is able to accurately estimate the fraction of baculoviral infected cells.
-
Cell infection can be detected in real time and earlier than offline methods.


Maarten Klaverdijk¹, Jort Altenburg¹, Dirk Martens¹, Gorben Pijlman², Jan van Hauwermeiren³, Jérémie Barbau³, Laurent Desmecht³, Damien Cabosart³
1. Bioprocess Engineering, Wageningen University and Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands,
2. Laboratory of Virology, Wageningen University and Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands,
3. Ovizio Imaging Systems, Rue du Bourdon 100/2 1180 Brussels, Belgium
Examination of vesicular stomatitis virus-induced morphology changes in individual Vero cells by qMod microscopy
Viral vectors are increasingly gaining importance in vaccine development, gene therapy and as oncolytic vectors. Vesicular stomatitis virus (VSV), an enveloped virus carrying a negative-sense RNA genome, has proven to be an excellent vaccine vector candidate against infectious diseases and specific cancers.
A variety of assays are available to determine yield of physical particles as well as infectious particles. However, several of the most well-characterized methods, such as plaque assays and tissue culture (TC) infectious dose 50 (TCID50) assays, are associated with a wide variance and are also time- and labor-intensive. Therefore, rapid and reliable means of assaying virus products are welcome advances, especially if the assay easily adapts to multiple product lines.
In this report, we describe a novel viral infection analysis method using a live VSV-based Lassa virus (LASV) vaccine candidate. The recombinant VSV has been genetically altered to express the LASV Josiah glycoprotein (VSVΔG/LASVGP), and infection in Vero cells was examined by microscopy using the Ovizio qMod camera and OsOne software. We assessed characteristics of infected Vero cells over the course of infection, obtained feature measurements of individual cells and examined very early biophysical distinctions of infected cells.
Process Development, Ology Bioservices, Alachua, FL 32615, USA
Isabel Scholz*, Christopher Montoya & Eric Vela
Digital holographic microscopy: a novel tool to study the morphology, physiology and ecology of diatoms.
Eva-Maria Zetsche, Ahmed El Mallahi & Filip J. R. Meysman. Diatom Research, 31:1, 1-16, 2016.
A recent publication by Zetsche et al. (2016) highlights the difficulties in imaging aquatic organisms such as diatoms: “Diatom cells are for the large part transparent, and since transparent substances or objects, by definition, do not absorb light in appreciable quantities when suspended in water, these entities are hard to discriminate and detect by microscopy techniques that rely on intensity information alone”.
“Piper (2011) suggested that interference-based contrast microscopy reveals the shape and structure of cells more clearly, as it improves the plasticity and contour sharpness.” Digital holographic microscopy (DHM) is in fact an interference-based approach and Dr. Zetsche and her co-authors are able to show that with DHM “the structural organization of diatoms is more clearly determined, in terms of cellular components, shapes and features.”
Ovizio’s qMod, a differential digital holographic microscopy camera for classical microscopes, is one of the instruments which offers a tool for the improved discrimination of living and dead diatoms (success rate >95%). Possible future applications can be the live-dead discrimination of microscopic aquatic organisms, as well as improved species identification. “Certain species of diatoms are frequently used to assess the water quality of rivers and lakes as well as coastal areas (Anton-Garrido et al. 2013, Kelly et al. 2009, Sabater et al. 2007). DHM may facilitate the live-dead differentiation of cells and thus improve these monitoring procedures” (Zetsche et al. 2016).

Figure: (a) Hologram of a cleaned frustule of Stauroneis sp. (University of Gent, Belgium) as obtained with an Ovizio digital holographic microscope. This hologram contains both light-intensity information (b) as well as phase information (c) representing the optical path length (OPL) of the object. (d) The OPL of an object is more clearly visualized with false coloring of the phase information. (Taken from Zetsche et al. 2016)